Tip
An interactive online version of this notebook is available, which can be
accessed via
Alternatively, you may download this notebook and run it offline.
Attention
You are viewing this notebook on the latest version of the documentation, where these notebooks may not be compatible with the stable release of PyBaMM since they can contain features that are not yet released. We recommend viewing these notebooks from the stable version of the documentation. To install the latest version of PyBaMM that is compatible with the latest notebooks, build PyBaMM from source.
Plot voltage components#
In a 1D model, the voltage of a cell is given by the difference in solid-phase potentials at the two edges of the domain
We can decompose this voltage into constituent components by adding and subtracting terms and regrouping:
where
and \(\bar{\cdot}\) represents the average value of the variable over its subdomain.
To demonstrate this, we solve a standard DFN model with the Chen2020 parameter set and plot the various potentials
[1]:
%pip install "pybamm[plot,cite]" -q # install PyBaMM if it is not installed
import pybamm
model = pybamm.lithium_ion.DFN()
experiment = pybamm.Experiment(["Discharge at 1C until 2.5 V"])
sim = pybamm.Simulation(
model, experiment=experiment, parameter_values=pybamm.ParameterValues("Chen2020")
)
sol = sim.solve()
sol.plot(
[
"Negative electrode bulk open-circuit potential [V]",
"Positive electrode bulk open-circuit potential [V]",
"Negative particle concentration overpotential [V]",
"Positive particle concentration overpotential [V]",
"X-averaged negative electrode reaction overpotential [V]",
"X-averaged positive electrode reaction overpotential [V]",
"X-averaged concentration overpotential [V]",
"X-averaged electrolyte ohmic losses [V]",
"X-averaged negative electrode ohmic losses [V]",
"X-averaged positive electrode ohmic losses [V]",
],
)
Note: you may need to restart the kernel to use updated packages.
At t = 472.93 and h = 1.932e-14, the corrector convergence failed repeatedly or with |h| = hmin.
At t = 232.93 and h = 3.07648e-13, the corrector convergence failed repeatedly or with |h| = hmin.
[1]:
<pybamm.plotting.quick_plot.QuickPlot at 0x1217ec4f0>
A more useful visualization is given by the plot_voltage_components function, which can either plot all the individual voltage components
[2]:
sol.plot_voltage_components(split_by_electrode=True)
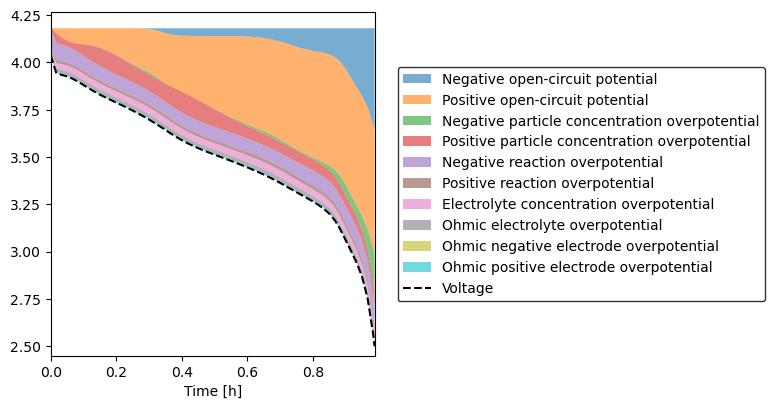
[2]:
(<Figure size 800x400 with 1 Axes>, <AxesSubplot: xlabel='Time [h]'>)
or group positive and negative together
[3]:
sol.plot_voltage_components()
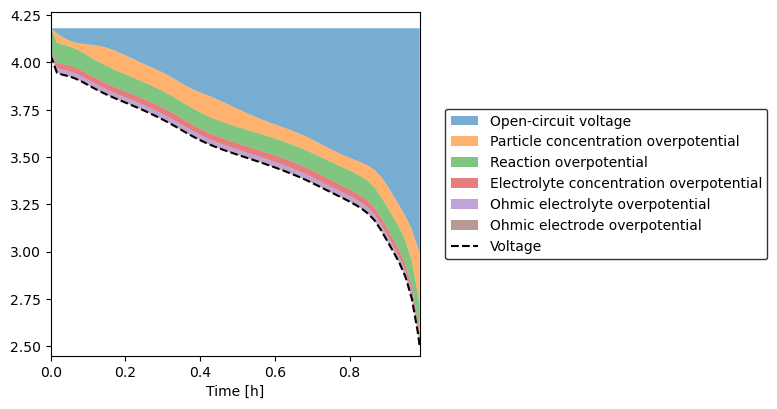
[3]:
(<Figure size 800x400 with 1 Axes>, <AxesSubplot: xlabel='Time [h]'>)
Note that this function further splits the electrolyte overpotential into the contribution from the electrolyte gradient and the contribution from ohmic losses
References#
The relevant papers for this notebook are:
[4]:
pybamm.print_citations()
[1] Weilong Ai, Ludwig Kraft, Johannes Sturm, Andreas Jossen, and Billy Wu. Electrochemical thermal-mechanical modelling of stress inhomogeneity in lithium-ion pouch cells. Journal of The Electrochemical Society, 167(1):013512, 2019. doi:10.1149/2.0122001JES.
[2] Joel A. E. Andersson, Joris Gillis, Greg Horn, James B. Rawlings, and Moritz Diehl. CasADi – A software framework for nonlinear optimization and optimal control. Mathematical Programming Computation, 11(1):1–36, 2019. doi:10.1007/s12532-018-0139-4.
[3] Chang-Hui Chen, Ferran Brosa Planella, Kieran O'Regan, Dominika Gastol, W. Dhammika Widanage, and Emma Kendrick. Development of Experimental Techniques for Parameterization of Multi-scale Lithium-ion Battery Models. Journal of The Electrochemical Society, 167(8):080534, 2020. doi:10.1149/1945-7111/ab9050.
[4] Rutooj Deshpande, Mark Verbrugge, Yang-Tse Cheng, John Wang, and Ping Liu. Battery cycle life prediction with coupled chemical degradation and fatigue mechanics. Journal of the Electrochemical Society, 159(10):A1730, 2012. doi:10.1149/2.049210jes.
[5] Marc Doyle, Thomas F. Fuller, and John Newman. Modeling of galvanostatic charge and discharge of the lithium/polymer/insertion cell. Journal of the Electrochemical society, 140(6):1526–1533, 1993. doi:10.1149/1.2221597.
[6] Charles R. Harris, K. Jarrod Millman, Stéfan J. van der Walt, Ralf Gommers, Pauli Virtanen, David Cournapeau, Eric Wieser, Julian Taylor, Sebastian Berg, Nathaniel J. Smith, and others. Array programming with NumPy. Nature, 585(7825):357–362, 2020. doi:10.1038/s41586-020-2649-2.
[7] Peyman Mohtat, Suhak Lee, Jason B Siegel, and Anna G Stefanopoulou. Towards better estimability of electrode-specific state of health: decoding the cell expansion. Journal of Power Sources, 427:101–111, 2019.
[8] Valentin Sulzer, Scott G. Marquis, Robert Timms, Martin Robinson, and S. Jon Chapman. Python Battery Mathematical Modelling (PyBaMM). Journal of Open Research Software, 9(1):14, 2021. doi:10.5334/jors.309.
[9] Pauli Virtanen, Ralf Gommers, Travis E. Oliphant, Matt Haberland, Tyler Reddy, David Cournapeau, Evgeni Burovski, Pearu Peterson, Warren Weckesser, Jonathan Bright, and others. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature Methods, 17(3):261–272, 2020. doi:10.1038/s41592-019-0686-2.
