Tip
An interactive online version of this notebook is available, which can be
accessed via
Alternatively, you may download this notebook and run it offline.
Attention
You are viewing this notebook on the latest version of the documentation, where these notebooks may not be compatible with the stable release of PyBaMM since they can contain features that are not yet released. We recommend viewing these notebooks from the stable version of the documentation. To install the latest version of PyBaMM that is compatible with the latest notebooks, build PyBaMM from source.
Compare lithium-ion battery models#
We compare three one-dimensional lithium-ion battery models: the Doyle-Fuller-Newman (DFN) model, the single particle model (SPM), and the single particle model with electrolyte (SPMe). Further details on these models can be found in [4].
Key steps:#
Comparing models consists of 6 easy steps:
Load models and geometry
Process parameters
Mesh the geometry
Discretise models
Solve models
Plot results
But, as always we first import pybamm and other required modules
[1]:
%pip install "pybamm[plot,cite]" -q # install PyBaMM if it is not installed
import pybamm
import os
os.chdir(pybamm.__path__[0] + "/..")
import numpy as np
import matplotlib.pyplot as plt
WARNING: You are using pip version 22.0.4; however, version 22.3.1 is available.
You should consider upgrading via the '/home/mrobins/git/PyBaMM/env/bin/python -m pip install --upgrade pip' command.
Note: you may need to restart the kernel to use updated packages.
1. Load models#
Since the three models we want to compare are already implemented in PyBaMM, they can be easy loaded using:
[2]:
dfn = pybamm.lithium_ion.DFN()
spme = pybamm.lithium_ion.SPMe()
spm = pybamm.lithium_ion.SPM()
To allow us to perform the same operations on each model easily, we create a dictionary of these three models:
[3]:
models = {"DFN": dfn, "SPM": spm, "SPMe": spme}
Each model can then be accessed using:
[4]:
models["DFN"]
[4]:
<pybamm.models.full_battery_models.lithium_ion.dfn.DFN at 0x7f917c709f40>
For each model, we must also provide a cell geometry. The geometry is different for different models; for example, the SPM has solves for a single particle in each electrode whereas the DFN solves for many particles. For simplicity, we use the default geometry associated with each model but note that this can be easily changed.
[5]:
geometry = {
"DFN": dfn.default_geometry,
"SPM": spm.default_geometry,
"SPMe": spme.default_geometry,
}
2. Process parameters#
For simplicity, we use the default parameters values associated with the DFN model, but change the current function to be an input so that we can quickly solve the model with different currents
[6]:
param = dfn.default_parameter_values
param["Current function [A]"] = "[input]"
It is simple to change this to a different parameter set if desired.
We then process the parameters in each of the models and geometries using this parameter set:
[7]:
for model_name in models.keys():
param.process_model(models[model_name])
param.process_geometry(geometry[model_name])
3. Mesh geometry#
We use the defaults mesh properties (the types of meshes and number of points to be used) for simplicity to generate a mesh of each model geometry. We store these meshes in a dictionary of similar structure to the geometry and models dictionaries:
[8]:
mesh = {}
for model_name, model in models.items():
mesh[model_name] = pybamm.Mesh(
geometry[model_name], model.default_submesh_types, model.default_var_pts
)
4. Discretise model#
We now discretise each model using its associated mesh and the default spatial method associated with the model:
[9]:
for model_name, model in models.items():
disc = pybamm.Discretisation(mesh[model_name], model.default_spatial_methods)
disc.process_model(model)
5. Solve model#
We now solve each model using the default solver associated with each model:
[10]:
timer = pybamm.Timer()
solutions = {}
t_eval = np.linspace(0, 3600, 300) # time in seconds
for model_name, model in models.items():
solver = pybamm.CasadiSolver()
timer.reset()
solution = solver.solve(model, t_eval, inputs={"Current function [A]": 1})
print(f"Solved the {model.name} in {timer.time()}")
solutions[model_name] = solution
Solved the Doyle-Fuller-Newman model in 266.815 ms
Solved the Single Particle Model in 20.571 ms
Solved the Single Particle Model with electrolyte in 38.459 ms
6. Plot results#
To plot results, we extract the variables from the solutions dictionary. Matplotlib can then be used to plot the voltage predictions of each models as follows:
[11]:
for model_name, model in models.items():
time = solutions[model_name]["Time [s]"].entries
voltage = solutions[model_name]["Voltage [V]"].entries
plt.plot(time, voltage, lw=2, label=model.name)
plt.xlabel("Time [s]", fontsize=15)
plt.ylabel("Voltage [V]", fontsize=15)
plt.legend(fontsize=15)
plt.show()
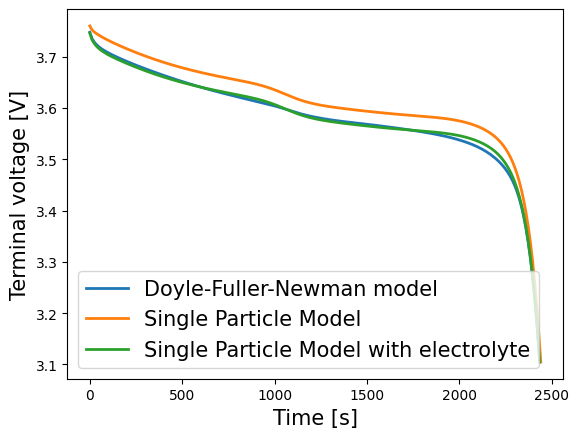
Alternatively the inbuilt QuickPlot functionality can be employed to compare a set of variables over the discharge. We must first create a list of the solutions
[12]:
list_of_solutions = list(solutions.values())
And then employ QuickPlot:
[13]:
quick_plot = pybamm.QuickPlot(list_of_solutions)
quick_plot.dynamic_plot();
Changing parameters#
Since we have made current an input, it is easy to change it and then perform the calculations again:
[14]:
# update parameter values and solve again
# simulate for shorter time
t_eval = np.linspace(0, 800, 300)
for model_name, model in models.items():
solutions[model_name] = model.default_solver.solve(
model, t_eval, inputs={"Current function [A]": 3}
)
# Plot
list_of_solutions = list(solutions.values())
quick_plot = pybamm.QuickPlot(list_of_solutions)
quick_plot.dynamic_plot();
By increasing the current we observe less agreement between the models, as expected.
References#
The relevant papers for this notebook are:
[15]:
pybamm.print_citations()
[1] Weilong Ai, Ludwig Kraft, Johannes Sturm, Andreas Jossen, and Billy Wu. Electrochemical thermal-mechanical modelling of stress inhomogeneity in lithium-ion pouch cells. Journal of The Electrochemical Society, 167(1):013512, 2019. doi:10.1149/2.0122001JES.
[2] Joel A. E. Andersson, Joris Gillis, Greg Horn, James B. Rawlings, and Moritz Diehl. CasADi – A software framework for nonlinear optimization and optimal control. Mathematical Programming Computation, 11(1):1–36, 2019. doi:10.1007/s12532-018-0139-4.
[3] Rutooj Deshpande, Mark Verbrugge, Yang-Tse Cheng, John Wang, and Ping Liu. Battery cycle life prediction with coupled chemical degradation and fatigue mechanics. Journal of the Electrochemical Society, 159(10):A1730, 2012. doi:10.1149/2.049210jes.
[4] Marc Doyle, Thomas F. Fuller, and John Newman. Modeling of galvanostatic charge and discharge of the lithium/polymer/insertion cell. Journal of the Electrochemical society, 140(6):1526–1533, 1993. doi:10.1149/1.2221597.
[5] Charles R. Harris, K. Jarrod Millman, Stéfan J. van der Walt, Ralf Gommers, Pauli Virtanen, David Cournapeau, Eric Wieser, Julian Taylor, Sebastian Berg, Nathaniel J. Smith, and others. Array programming with NumPy. Nature, 585(7825):357–362, 2020. doi:10.1038/s41586-020-2649-2.
[6] Scott G. Marquis, Valentin Sulzer, Robert Timms, Colin P. Please, and S. Jon Chapman. An asymptotic derivation of a single particle model with electrolyte. Journal of The Electrochemical Society, 166(15):A3693–A3706, 2019. doi:10.1149/2.0341915jes.
[7] Valentin Sulzer, Scott G. Marquis, Robert Timms, Martin Robinson, and S. Jon Chapman. Python Battery Mathematical Modelling (PyBaMM). Journal of Open Research Software, 9(1):14, 2021. doi:10.5334/jors.309.
